Technology Overview
Eclipsebio has optimized eCLIP (enhanced Crosslinking and Immunoprecipitation sequencing) developed at UCSD by incorporating modifications to the workflow that improve the accuracy and sensitivity of the results. RBP-eCLIP™ is a powerful tool to investigate protein-RNA interactions on a transcriptome-wide scale.
RNA binding proteins (RBPs) bind to RNAs through recognition of sequence and structural motifs to regulate RNA function in a cell-type, condition-specific, or temporal manner. Recent studies have estimated that there are over 1500 RBPs in the human genome and these RBPs play an integral role modulating RNA stability and function throughout the RNA life cycle. RBPs play crucial roles in post-transcriptional gene regulation, including mRNA splicing, stability, localization, and translation. Mutations in RBPs have been linked to cancer, neurodegenerative diseases, muscular dystrophies, cardiac diseases, metabolic disorders, autoimmune diseases and numerous others.
RBP-eCLIP™ has broad applications in understanding fundamental biological processes such as development, differentiation, and disease. It can shed light on the regulatory networks involved in various cellular processes and help identify novel RNA-based biomarkers and therapeutic targets.
Additionally, RBP-eCLIP provides insights into RNA-binding protein mutations associated with human diseases and contributes to our understanding of RNA-based regulatory mechanisms. Researchers can utilize these datasets to study specific RBPs or perform integrative analyses with other genomic data, facilitating the generation of comprehensive and multi-dimensional models of gene regulation.
Need inspiration for your next RBP-eCLIP project? Interact with analyzed results from over 150 RBPs with complimentary ENCODE datasets by Eclipsebio. All reports contain data for peaks and motif information specific for your RBP of interest. Visualize data your way and compare your own results to ENCODE.
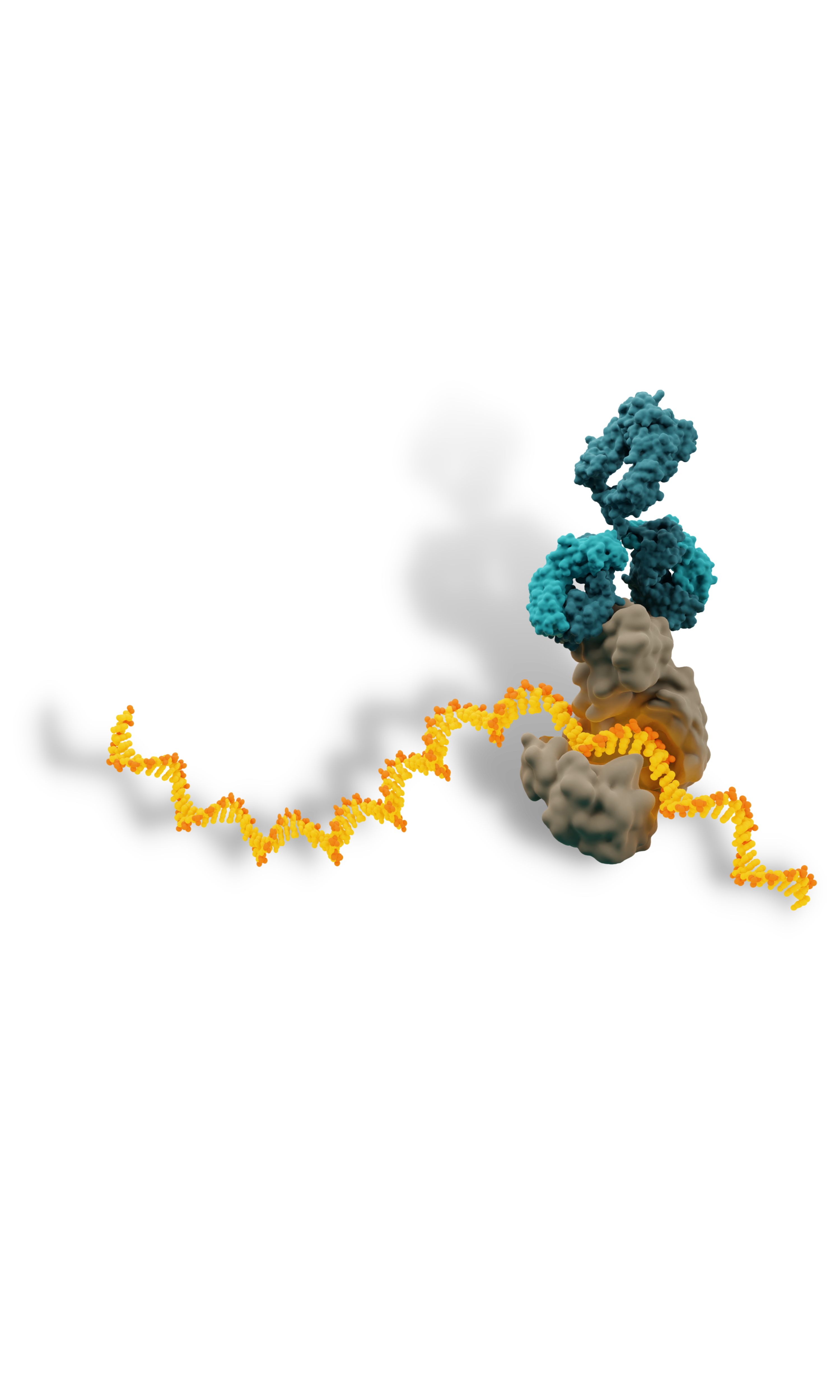
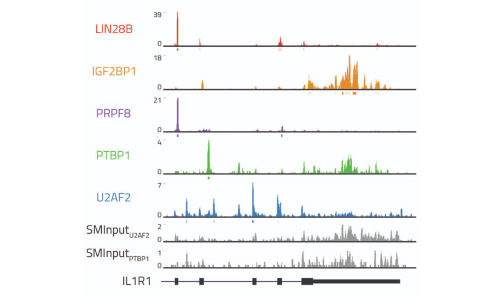
Transcriptome-wide Identification of RNA Targets
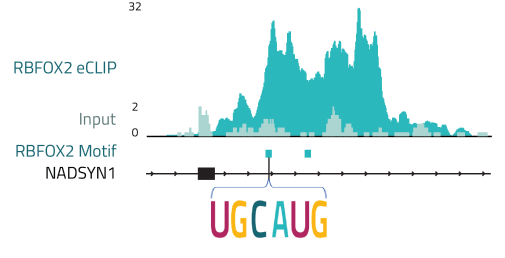
Enabling RBP RNA binding motif identification
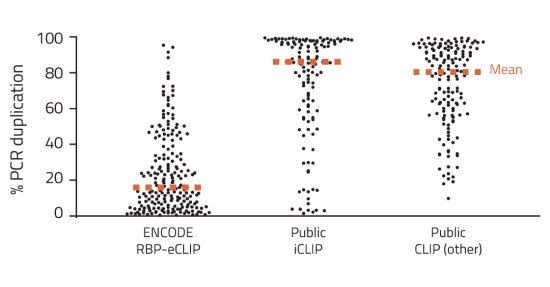
High Efficiency Library Preparation with Decreased PCR Duplication
- 1000-fold increase in library efficiency compared to other CLIP methods
- RBP-eCLIP increases experimental success rates and decreases wasted sequencing due to PCR duplication